Illuminating compartmentalized metabolism
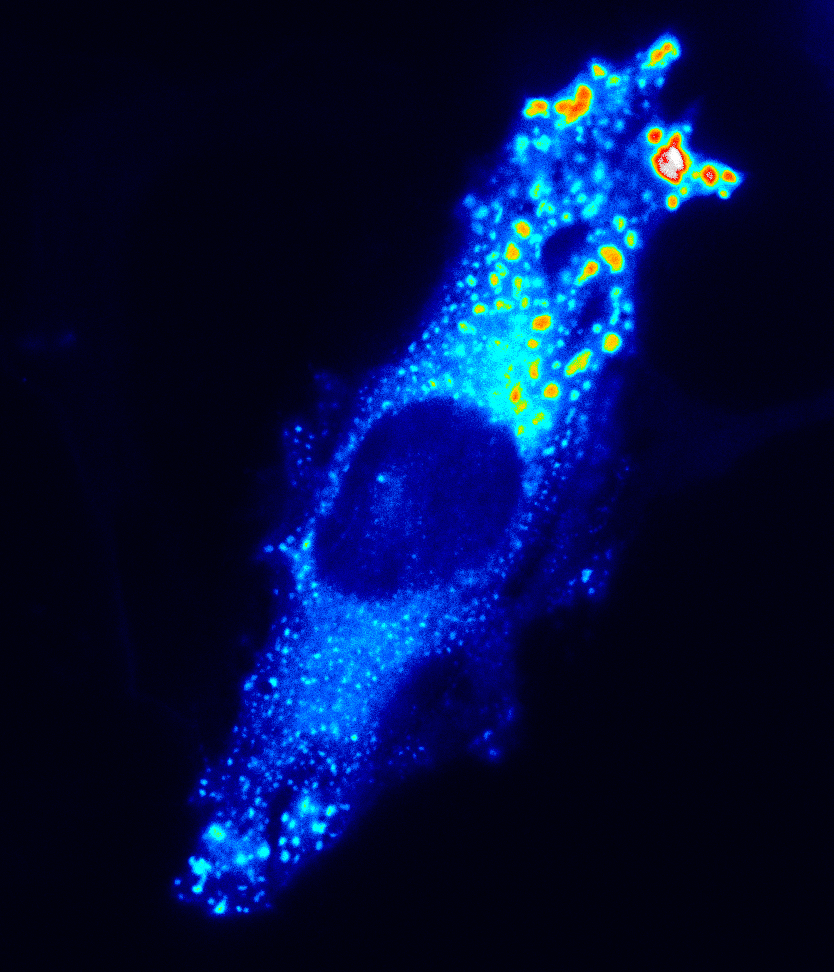
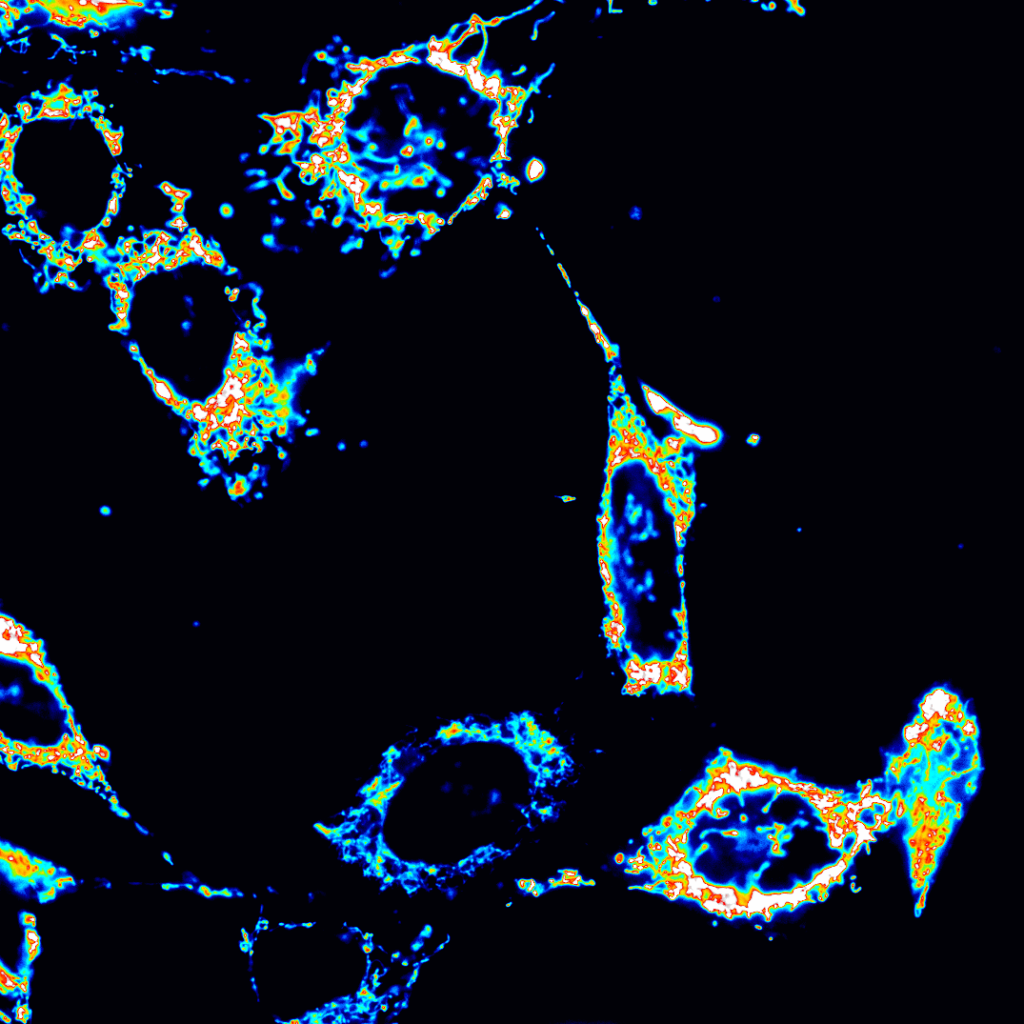
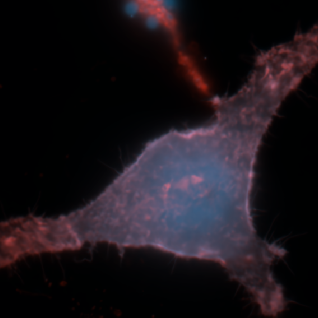
The cell is a highly complex environment with numerous processes occurring in a micron-sized environment. How does the cell organize essential processes like metabolism and signaling to coordinate function and fate?
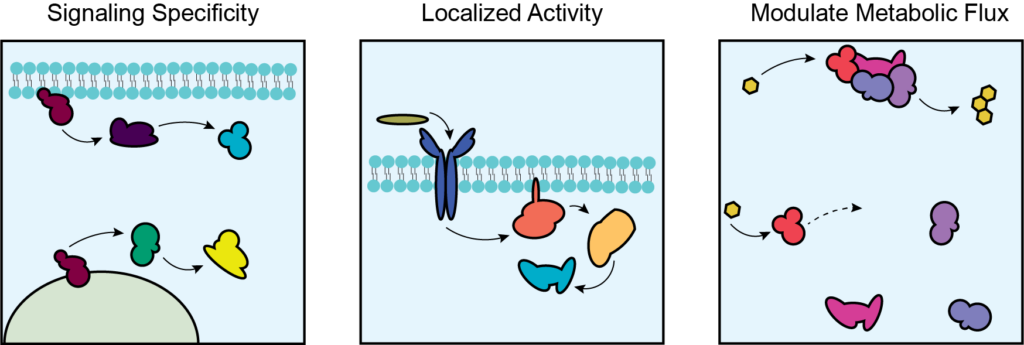
The cell closely coordinates processes through compartmentation, allowing for signal specificity, localized activity, and modulation of metabolic flux. But how does this compartmentation occur? What are mechanisms for compartmentation of metabolic processes? How does metabolic disease perturb compartmentation?
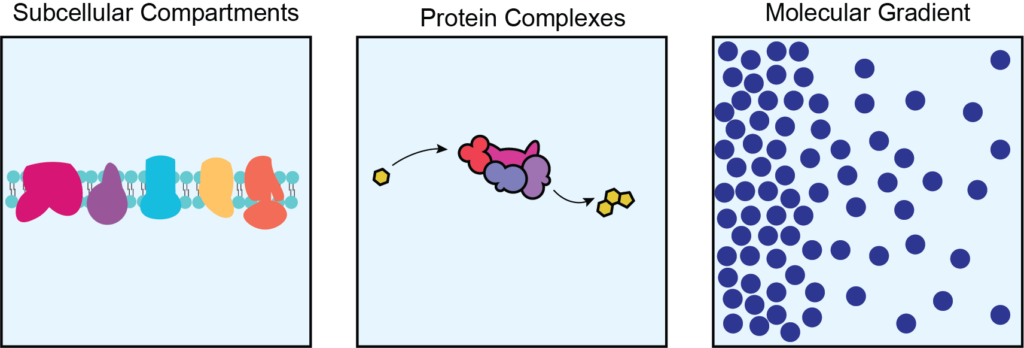
Organization of metabolic proteins, metabolites, and related signaling networks can be achieved through compartmentalization of cellular processes in or around organelles, within protein complexes, and in molecular gradients to provide specificity and fine-tuning of cell activities. The Schmitt Lab focuses on identifying mechanisms for subcellular compartmentation of metabolic processes, including signaling networks regulating metabolism and metabolite compartmentation.
The Schmitt Lab develops genetically encoded fluorescence microscopy-based tools to study cellular activity in single cells in real time. These molecular tools provide high spatial and temporal resolution of cellular behavior.
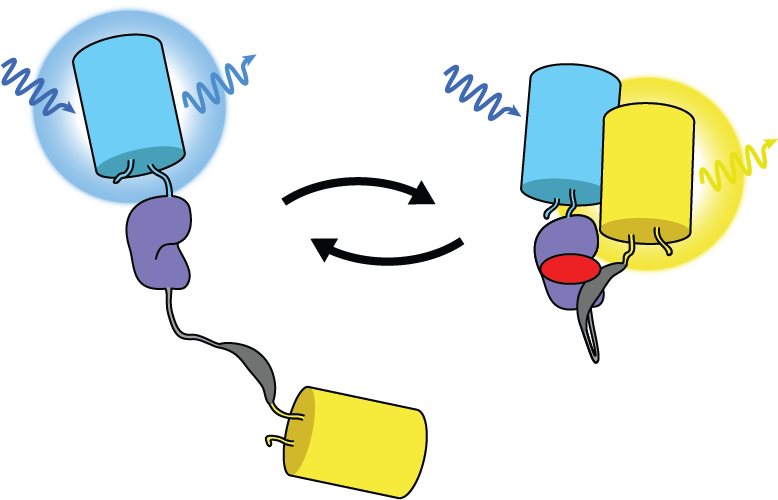
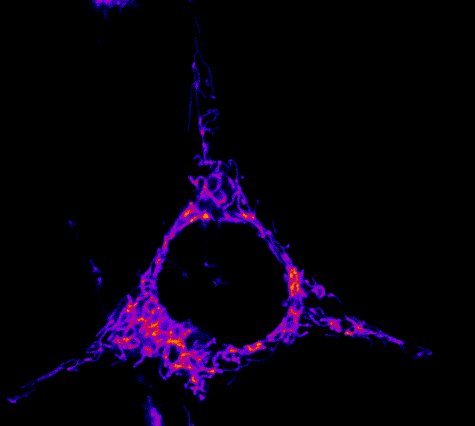
We develop and use biosensors for quantitative imaging to (1) study kinase signaling networks which regulate metabolic processes and (2) interrogate flux of molecules like amino acids and metabolites in real time in single cells. We can study dynamic biochemical processes in real-time, at the single-cell level, with high spatial and temporal resolution, thus illuminating biochemistry in native environments.
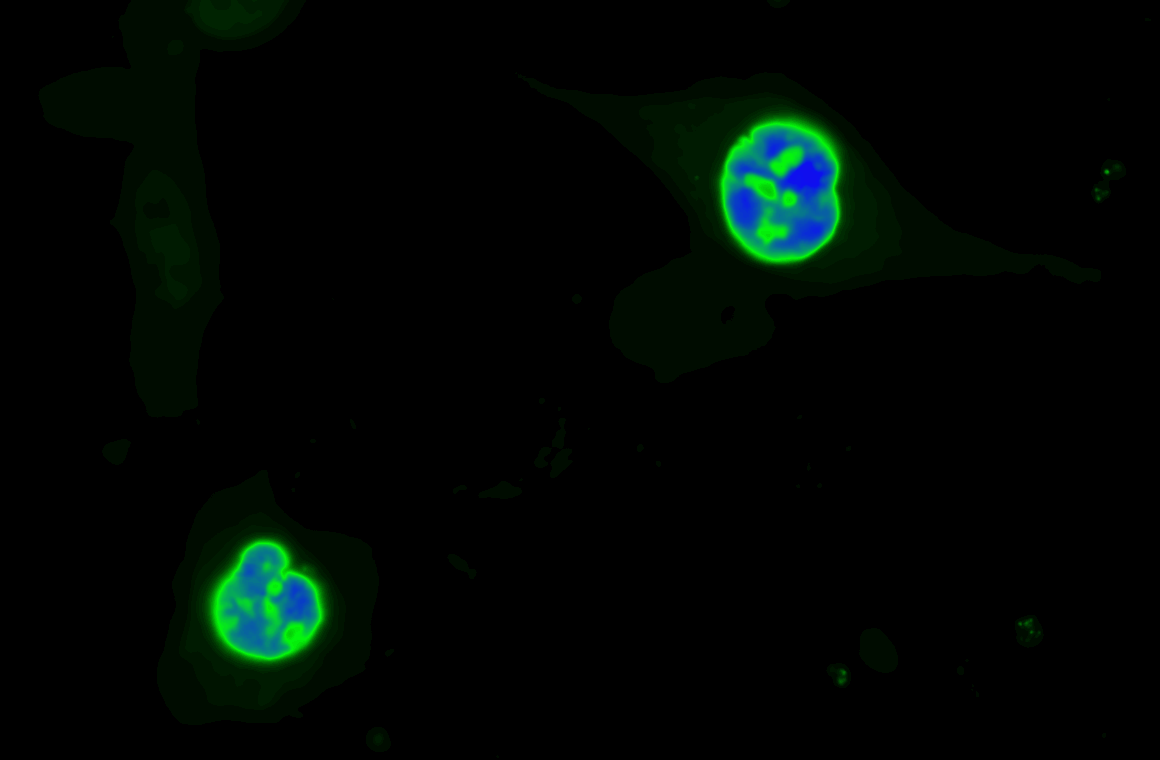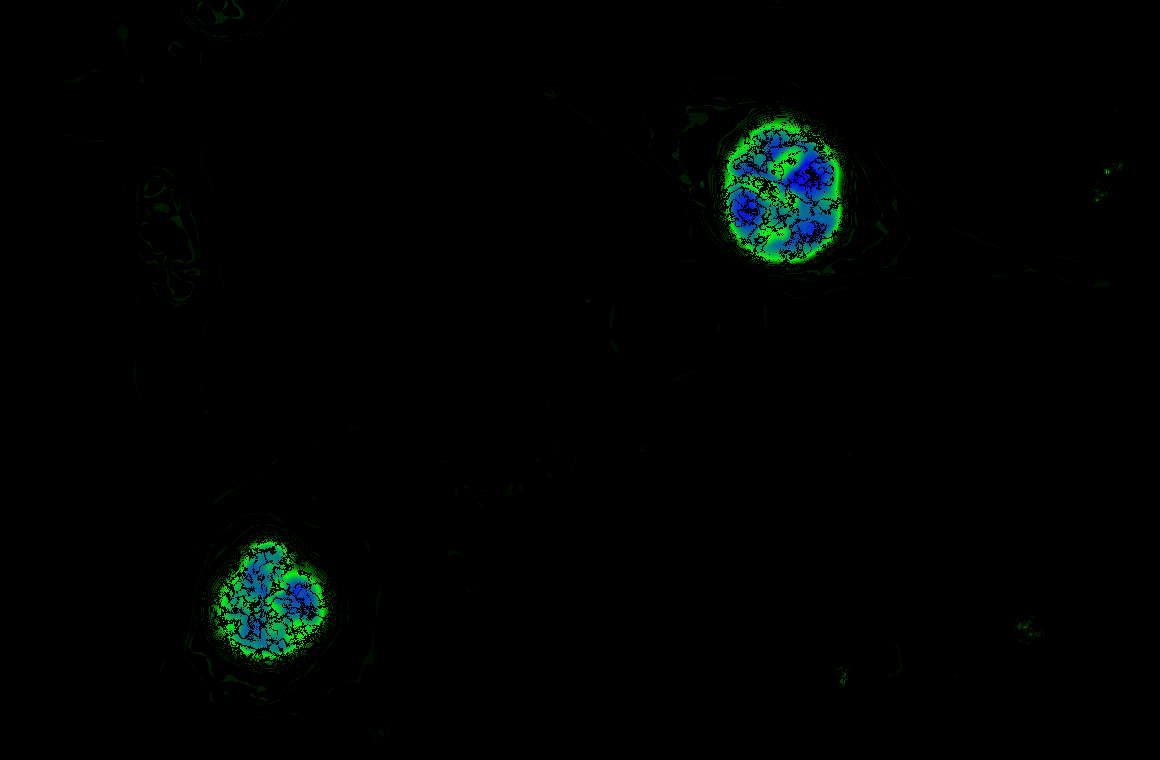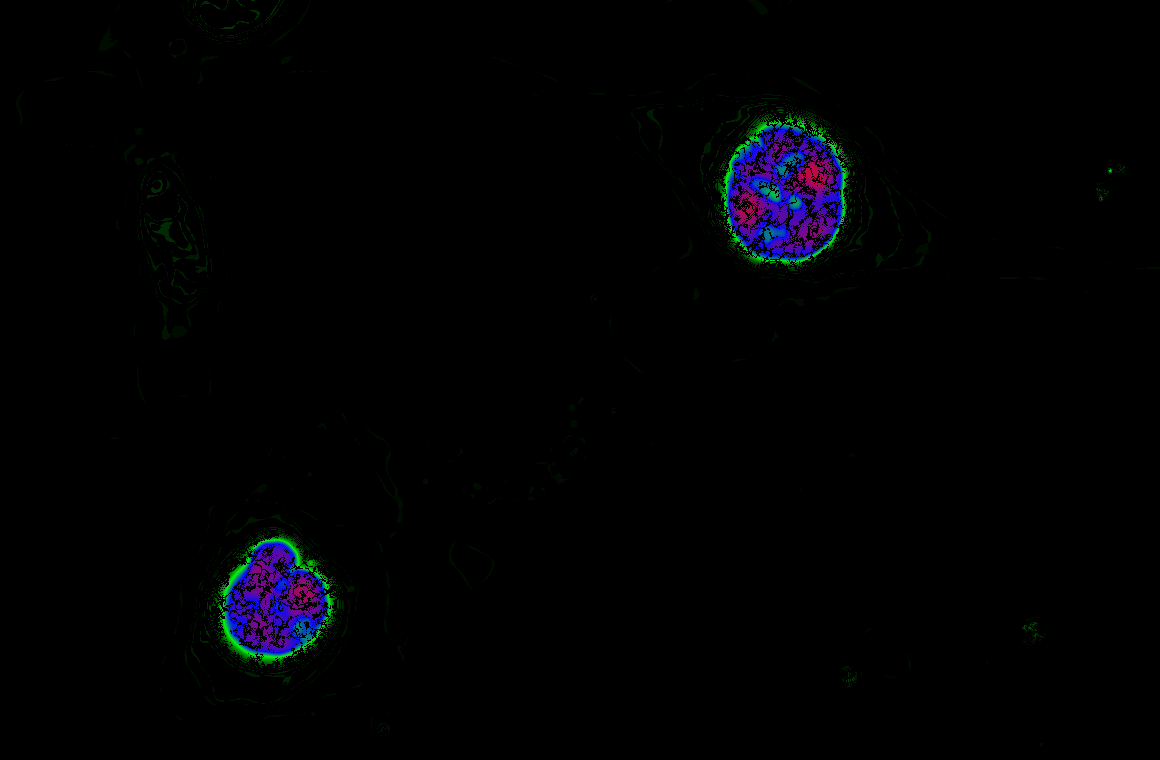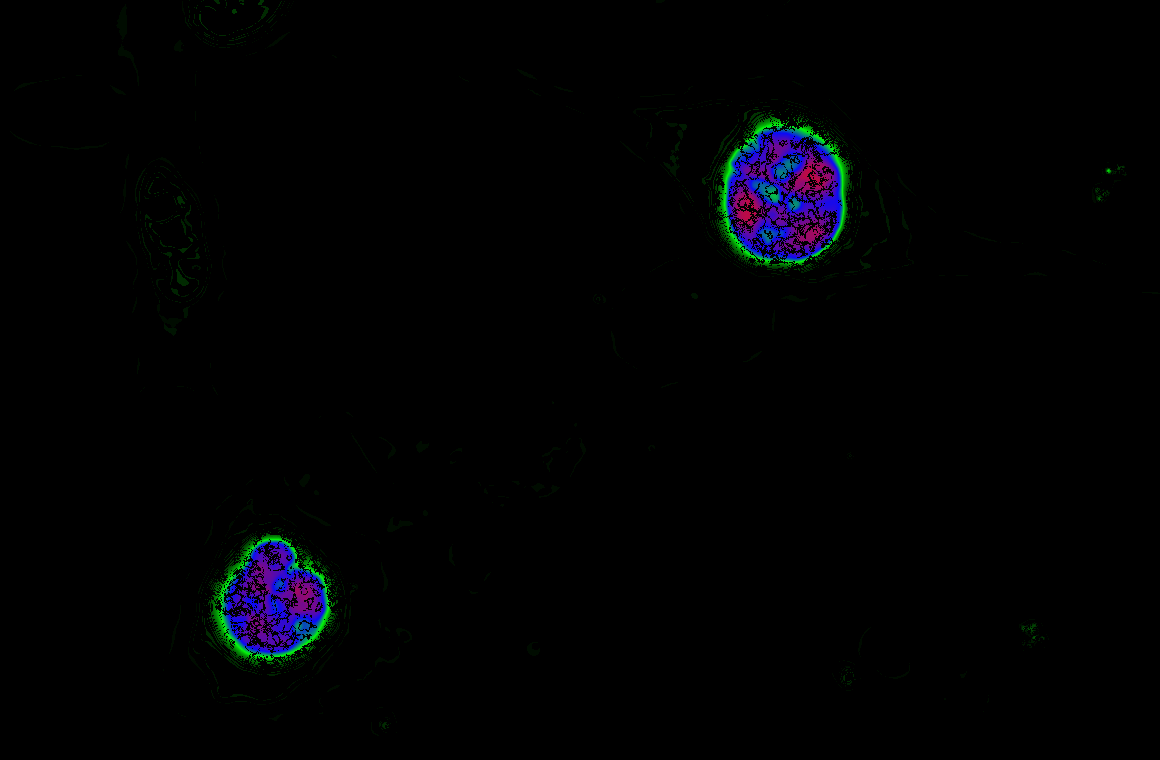
We focus on answering questions surrounding how compartmentation of metabolic processes is achieved, and how metabolic diseases, like cancer and inborn errors of metabolism, perturb this compartmentation.